
Sleeper cells: New neural stem cell state gives insight into cancer
ASU researchers discover and explore new resting phase of stem cells that could help put brain tumors to sleep

Above: Arizona State University biomedical engineering researchers developed a new cell classifier tool that takes a higher-resolution look at the life cycle of neuroepithelial stem cells, which led to the discovery and exploration of a new resting phase called Neural G0. This knowledge could help scientists to better understand glioma brain tumors and develop new methods of treatment. Graphic by Rhonda Hitchcock-Mast and Christopher Plaisier
The human body contains approximately 200 different kinds of cells, including many kinds of stem cells that have the potential to turn into a variety of specialized cells. Probing into the life cycle states or phases of these cells is important to understanding and treating diseases like dementia and cancer.
Christopher Plaisier, an assistant professor of biomedical engineering in the Ira A. Fulton Schools of Engineering at Arizona State University, and Samantha O’Connor, a biomedical engineering doctoral student in the Plaisier Lab, are leading research into a new state of the stem cell life cycle that could be the key to unlocking new methods of brain cancer treatment. Their work was recently published in the research journal Molecular Systems Biology.
“The cell cycle is such a well-studied thing and yet here we are looking at it again for the umpteenth time and a new phase pops out at us,” Plaisier says. “Biology always has new insights to show us, you just have to look.”
Taking a closer look
The spark for this discovery came through a collaboration with Patrick Paddison, an associate professor at the Fred Hutchinson Cancer Research Center in Seattle, and Dr. Anoop Patel, an assistant professor of neurological surgery at the University of Washington who is also involved in the Fred Hutchinson Cancer Research Center. Paddison’s team was looking at genes that play a role in stem cells’ growth in the brain as a potential path to a treatment for neurodegeneration.
“The primary feature of any cancer is that the cells are proliferating,” Plaisier says. “If we could get in there and figure out what the mechanisms are, that might be a place to slow them down.”
“In cases where you have neurodegeneration, having stem cells that proliferate is potentially beneficial,” Plaisier says. “The one problem with that is it’s also similar to what happens when cancer cells increase rapidly. It’s two sides of the same coin.”
Paddison’s team called upon Plaisier to help analyze their brain stem cell data characterized through a process called single-cell RNA sequencing.
“That data turned out to be pretty amazing,” Plaisier says. “It mapped out into this beautiful circular pattern that we identified as all of the different phases of the cell cycle.”
By taking a closer look at the data, they found a new cell phase that had never been observed before in neuroepithelial stem cells, which they call Neural G0. This discovery sparked a six-year research project to study the new phase and determine what it means for the progression of diseases, particularly glioma brain tumors.
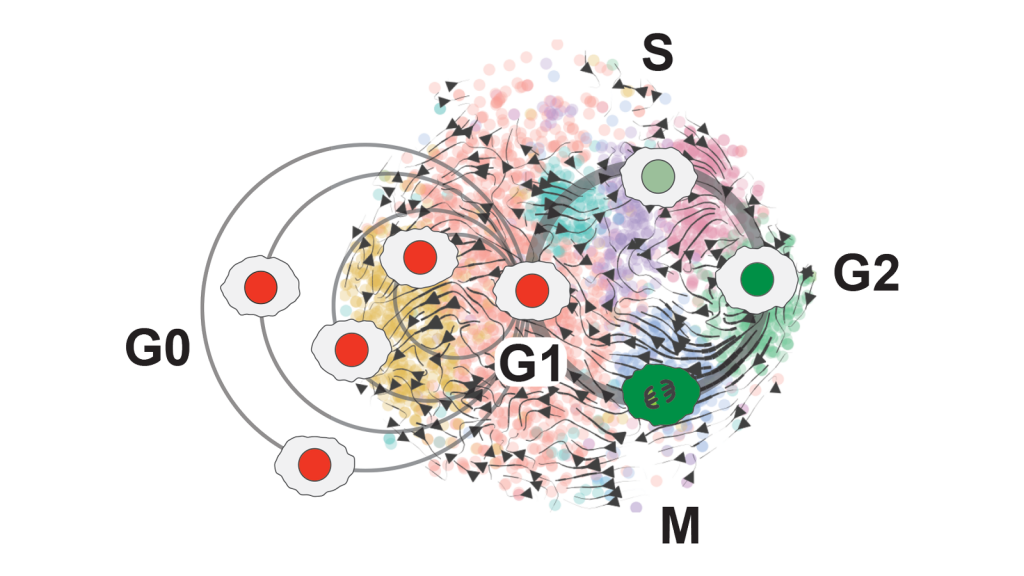
By analyzing brain stem cell data, Assistant Professor Christopher Plaisier and biomedical engineering doctoral student Samantha O’Connor saw the phases of the cell cycle mapped out in more detail than previously possible — the G0 resting phases, including a new, separate phase they called Neural G0; the first growth phase, G1; the genetic material copying phase, S; the second growth phase, G2; and the splitting phase, M. In addition to Neural G0, they also found new phases at the beginning and end of the cell cycle. Graphic courtesy of Christopher Plaisier
Understanding the life of a cell
Our body’s cells go through four main phases: grow, copy the genetic material stored in the nucleus, grow again and divide into two new cells. These phases are known as G1, S, G2 and M.
Many cells stop after their initial growth and enter an alternate state after G1, which is a sleep-like state called G0. In this state, cells are dormant or go about their normal functions without preparing to divide again.
Cancer arises when cells develop mutations that cause them to break out of the G0 state and grow and divide very quickly.
Like other types of cells, stem cells also have resting states. One is known as quiescence, which is the state where cells live independently from the cell cycle, though they can hop back into the cell cycle given the right cues. Neural G0 is similar to this quiescent state that other stem cells go through, but it has features particular to neuroepithelial cells, a type of stem cell that is destined to turn into certain brain cells.
Glioma tumors arise in the brain from neuroepithelial cells entering the growth and division phase at higher rates. Studying the life cycle of these cells is an important area of research for finding ways to combat cancer cell proliferation.
Developing new tools
Researchers use tools called classifiers to assign a cell cycle phase to individual cells based on the state of RNA messenger substances inside the cell.
Many existing cell cycle classifiers pick out only the major steps in a cell’s life cycle, like a map of the world that depicts only outlines of the continents. However, just as we know there are far more details that can be mapped on a continent, it turns out there is much more to see within the individual stages of the cell cycle.
“We were able to pick out phases that are glommed together in the other cell cycle classifiers,” Plaisier says. “That potentially has several different uses.”
The tool O’Connor developed — called ccAF, or cell cycle ASU/Fred Hutchinson to represent the collaboration between the two institutions — takes a closer, “high-resolution” look at what’s happening within the growth cycles of stem cells and identifies genes that can be used to track progress through the cell cycle.
“Our classifier gets deeper into the cell cycle because there could be pieces we’re capturing that have important implications for disease,” O’Connor says.
While it is possible to pick out the cell phases using complicated techniques such as applying fluorescent markers on actual cells, Plaisier and O’Connor found the Neural G0 state purely through looking at data, a much simpler process.
Plaisier and O’Connor are making the ccAF classifier tool open source and available in a variety of formats for anyone studying single-cell RNA sequencing data to ease into the process of studying cell cycles.
The tool could also be used to study other types of stem cells that potentially have yet-to-be-discovered G0 states similar to or completely different from the Neural G0 state of neuroepithelial stem cells.

Assistant Professor Christopher Plaisier is a cross-disciplinary researcher who integrates large-scale data in the study of and modeling of biological systems to help answer questions about human diseases. He and his research team, including doctoral student Samantha O’Connor, used an integrative computational systems biology approach to develop a new cell classifier tool to explore the newly discovered Neural G0 phase of stem cells in the brain. Photographer: Marco-Alexis Chaira/ASU
Applying new knowledge to study cancer
When Plaisier and O’Connor used the ccAF tool to analyze cell data for glioma tumors, they found the tumor cells were often either in the Neural G0 or G1 growth state. And as tumors become more aggressive, fewer and fewer cells remain in the resting Neural G0 state. This means more and more cells are proliferating and growing the tumor.
They correlated this data with the prognosis for patients with glioblastoma, a particularly aggressive type of brain tumor. Those with higher Neural G0 levels in tumor cells had less aggressive tumors.
They also found that the quiescent Neural G0 state is independent of a tumor’s proliferation rate, or how fast its cells divide and create new cells.
“That was an interesting finding from our results, that quiescence itself could be a different biological process,” Plaisier says. “It’s also a potential point where we could look for new drug treatments. If we could push more cells into that quiescent state, the tumors would become less aggressive.”
Current cancer drug treatments focus on killing cancer cells. However, when the cancer cells are killed, they release cell debris into the surrounding area of the tumor, which can cause the remaining cells to become more resistant to the drugs.
“So, instead of killing the cells, if we put them to sleep it could potentially be a much better situation,” Plaisier says.
For example, if a glioblastoma tumor is cut out, a drug that promotes the quiescent state could be introduced to the tumor site to prevent any remaining cells from growing and causing a tumor to recur.
“You can maintain what’s there and it wouldn’t get worse,” O’Connor says. “It’s a different idea of how to treat cancer.”
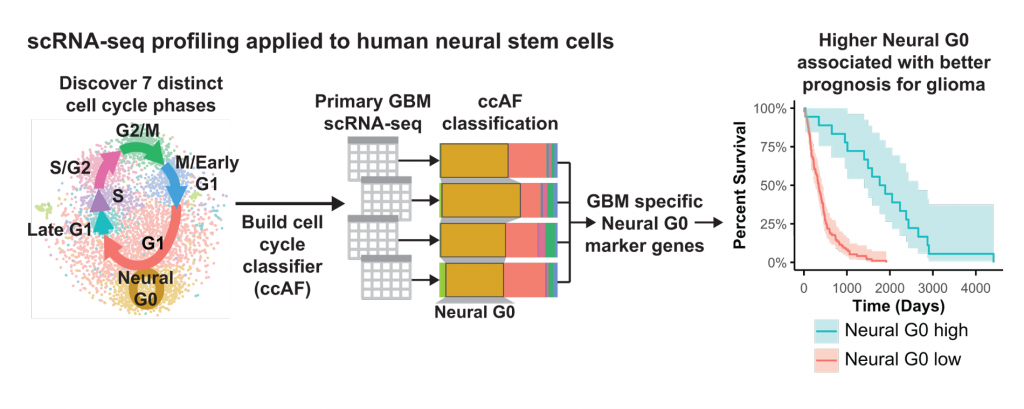
Christopher Plaisier, Samantha O’Connor and their research team used single-cell RNA sequencing, or scRNA-seq to discover seven distinct cell cycle phases, including the new Neural G0 resting phase. Plaisier and O’Connor built a new cell classifier tool called ccAF to further explore this phase. They also explored Neural G0’s relationship to glioblastoma, or GBM, cancer prognosis. They found that higher rates of Neural G0 cells were associated with better survival in patients with glioma tumors. Graphic courtesy of Christopher Plaisier
Further exploration
With the possibility of new cancer treatments in mind, Plaisier and O’Connor are exploring which genes are responsible for pushing cells into a quiescent state and how this function can be used for new therapies for glioblastomas, the most common and deadly brain tumor.
This research is part of a new project supported by a National Institutes of Health R01 grant to better define G0-like states in glioblastoma brain tumors. This NIH grant from the National Institute for Neurological Disorders and Stroke supports the cross-disciplinary collaboration between the Plaisier, Paddison and Patel labs to create a map of gene interactions that govern tumor regulation of G0-like states and explain therapy resistance and glioblastoma recurrence.
With their ccAF tool, they were also able to find new states at the beginning and end of the cell cycle that exist between the commonly known states.
“We’re starting to think about ways to dig into those and learn more about the biology of the entry and exit from the cell cycle because those are potentially really important points where the cells will either go into the G1 state or G0,” Plaisier says.
Figuring out what triggers a cell to enter the division cycle or remain in a G0 resting state could help understand the processes behind tumor growth.
“The primary feature of any cancer is that the cells are proliferating,” Plaisier says. “If we could get in there and figure out what the mechanisms are, that might be a place to slow them down.”
Cross-disciplinary skills lead to innovation
When biomedical engineering doctoral student Samantha O’Connor joined Assistant Professor Christopher Plaisier’s lab in 2019, she had plenty of experience culturing cells and conducting experiments in the lab, but it was the first time she was called upon to code a classifier and work on the computational side of biomedical engineering research.
“In a lot of cases, the people who collect the data or do the experiments will send it off to another person who does the analysis, but there’s no real connection to what the purpose of the study is,” O’Connor says. “To be able to do both really improves the output because you know what you want to find.”
This type of “integrative computational systems biology” is Plaisier’s focus.
“I take very big biological data sets and put them together in ways that help build models to make sense of biology and disease,” he says.
Plaisier worked as a software engineer between earning his bachelor’s degree in biology and starting graduate school to study human genetics and bioinformatics. His focus as a graduate student was bringing together biology and computation, and he has carried that theme into his work in the Fulton Schools.
“What I’m trying to do in the lab is train cross-disciplinary people,” Plaisier says. “Sam is learning how to program and take the data we’re generating and turn it into models and ideas and build things from that.”



































