ASU researcher chips away at disease diagnosis limitations
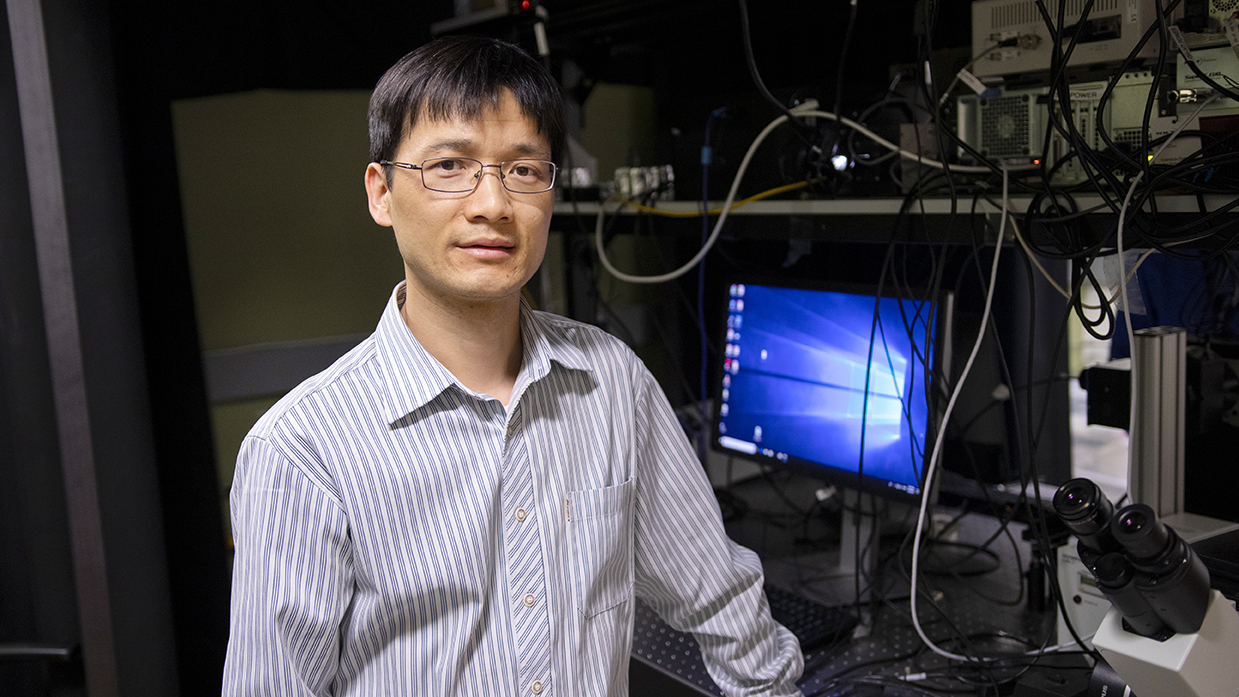
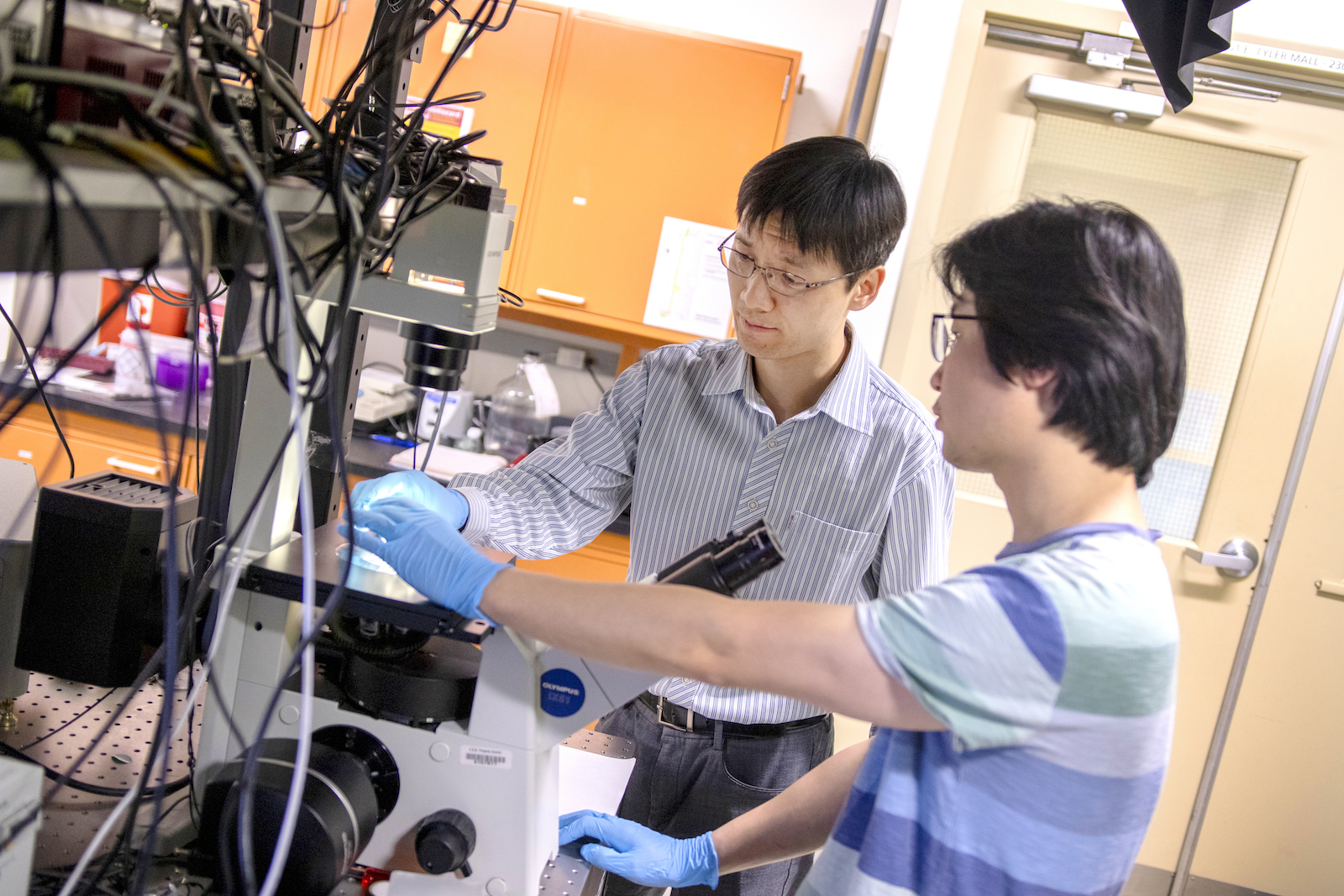
Above: Assistant Professor Chao Wang is one of 10 Ira A. Fulton Schools of Engineering faculty members to receive an National Science Foundation CAREER Award for 2018 to 2019. He is developing ExoMiRChip, an integrated fluidic device on a silicon chip to improve the capabilities of liquid biopsy to diagnose cancer and infectious and neurodegenerative diseases.
Ten faculty members in the Ira A. Fulton Schools of Engineering have received NSF CAREER Awards between September 2018 and April 2019.
Detecting a disease like cancer early often yields the best results — time is on your side for treatment.
Liquid biopsy is a non-invasive way to detect microscopic particles that can indicate the presence of a disease in its early stages. Current liquid biopsy methods, however, are time-consuming and frequently error-prone.
Chao Wang is working on a way to offer more time and certainty.
ExoMiRChip is a compact, integrated fluidic device on a silicon chip Wang is developing to make liquid biopsy faster, more sensitive and less invasive by using smaller blood samples. The assistant professor of electrical engineering in Arizona State University’s Ira A. Fulton Schools of Engineering has recently received a five-year, $500,000 National Science Foundation Faculty Early Career Development Program (CAREER) Award to support this work.
Exomes, a type of extracellular nanoparticle, are a valuable biomarker. Biomarkers are a measurable indicator of the presence or severity of cancers and infectious and neurodegenerative diseases.
ExoMiRChip uses microfluid physics to purify, or separate, samples of exomes and to quantify the exome samples for better diagnostic results.
“Because both the purification and sensing are completely compatible with silicon-based manufacturing, we are able to integrate the whole system on one platform,” Wang says. “Such an integration is advantageous because it potentially allows full-spectrum exome-based biomarker analysis from whole blood with minimized human intervention and sample contamination.”
To purify the exomes, a blood sample is run across the silicon chip’s circular and triangular nanostructures about one-thousandth of a human hair. The small exome particles move past the nanostructures in a zigzag fashion in one direction while large exome particles zigzag in the other direction.
ExoMiRChip then uses micro RNA sensors to detect the short nucleic acid molecules (micro RNA) of the exomes from each sized population. Properties of the micro RNA can then help identify different disease factors.
Through this two-step, integrated process, ExoMiRChip can reduce the speed of diagnosis from days or weeks to hours.
A unique aspect of ExoMiRChip’s methodology is that it increases analysis capabilities by using an entire exosome size population from the blood sample rather than just a small portion of the sample, which is typical of other testing methods. This technique also allows using blood samples that are only microliters in volume rather than milliliters.
“We can simultaneously analyze exosomes of different sizes, compared to other technologies that have to choose a specific size and have to discard the rest, which could cause biases in exosome analysis given the broad exosome size distribution and complexity in its biogenesis,” Wang says.
Wang, a faculty member in the School of Electrical, Computer and Energy Engineering, one of the six Fulton Schools, has been combining nanotechnology and biotechnology research for the past two years to accumulate preliminary data for the ExoMiRChip device. He believes bridging engineering and biology helped demonstrate to the NSF that this is a viable, high-impact project for the early stage diagnosis, prognosis and management of a wide range of diseases.
“The combined purification and sensing strategy will apply to the detection of other molecules other than micro RNA such as cell-free DNA, RNA and proteins,” Wang says. “Further, the integration [on a chip] will eventually enable the analysis of a panel of biomarkers, which will greatly improve diagnostic accuracy.”